The RNA infrastructure - RNA networks in the cell
RNA is well known as the transient molecule between the code in DNA and the proteins that eventuate from it. However, it is now known that many genes do not go all the way to the protein stage and function as non-coding RNAs (ncRNAs). There are many classes of ncRNAs in the RNA Underworld. There are ribosomal RNAs that make up the framework (the ribosome) upon which proteins are made, and transfer RNAs that deliver amino acids to the ribosome to construct the proteins. There is also a class of very small RNAs (~20 nucleotides) that regulate other genes in a process called RNA interference (RNAi), and very long ncRNAs (>200 nucleotides) that we are finding have functions in organism development. We have miRNA, siRNA, piRNA, qiRNA, rasiRNA…so many RNA classes in fact we may soon run out of letters to put in front!
All these RNA classes function differently, and have to be analysed in a different way from protein genes. This is because the RNA strand folds into a 3D structure then binds to specific proteins, meaning that analysis goes much further than looking at the sequence. My research examines the way these RNA genes network together in the cell and I have been finding that the levels of complexity for this networking have been very much understated. Within our cells we have an ‘underworld’ of RNA-based interactions which we call the RNA-infrastructure. This theme has come from the early idea that ncRNA was merely ‘junk’, but now is seen to be critical to cellular function. We now call ncRNA the ‘dark matter’ of the cell. The RNA Underworld is often understated but we need it to be there. In beginning this research I opened a real ‘pandora’s box’ and I do not want to close the lid; the contents are just too fascinating!
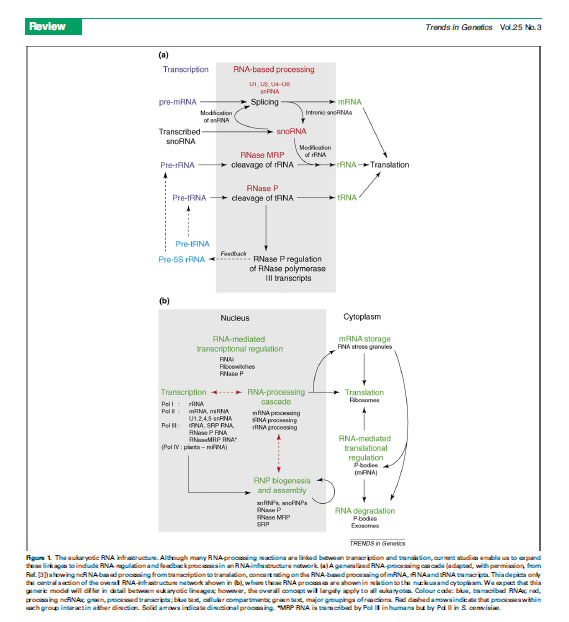
Figure from Collins and Penny 2005 The RNA Infrastructure: Dark Matter of the Eukaryotic Cell. Trends in Genetics 25 (3)120-128